Google Cloud-scale computation-driven drug discovery: Atommap computational drug discovery company built an elastic supercomputing cluster on Google Cloud for large-scale computation-driven drug discovery.
New medicine introduction usually involves four steps: Target identification locates the protein target linked to the illness; molecular discovery finds the novel molecule that modulates the target’s function; clinical trial evaluates the candidate drug molecule’s safety and efficacy in patients; and commercialization provides the medication to patients.
First, an efficient mechanism to modulate the target function must be established to maximize therapeutic efficacy and minimize side effects. Second, the ideal drug molecule must be designed, chosen, and manufactured to faithfully implement the mechanism, be bioavailable, and have acceptable toxicity.
Why is molecular discovery so difficult?
A protein is always moving due to heat, which modifies its conformation and its ability to connect with other biomolecules, ultimately impacting its functions. Time and again, structural information about a protein’s conformational dynamics points to new avenues for functional regulation. Nevertheless, despite recent enormous advancements in experimental methodologies, such information frequently eludes experimental determination.
The number of different tiny molecules in the chemical “universe” is estimated to be 1060 (Reymond et al., 2010). With perhaps ten billion already manufactured, chemists have roughly 1060 still to go.
Molecular discovery presents two primary obstacles despite its boundless potential: it’s likely that they haven’t explored every mechanism of action or identified the optimal molecules, meaning the could always create a more effective medication.
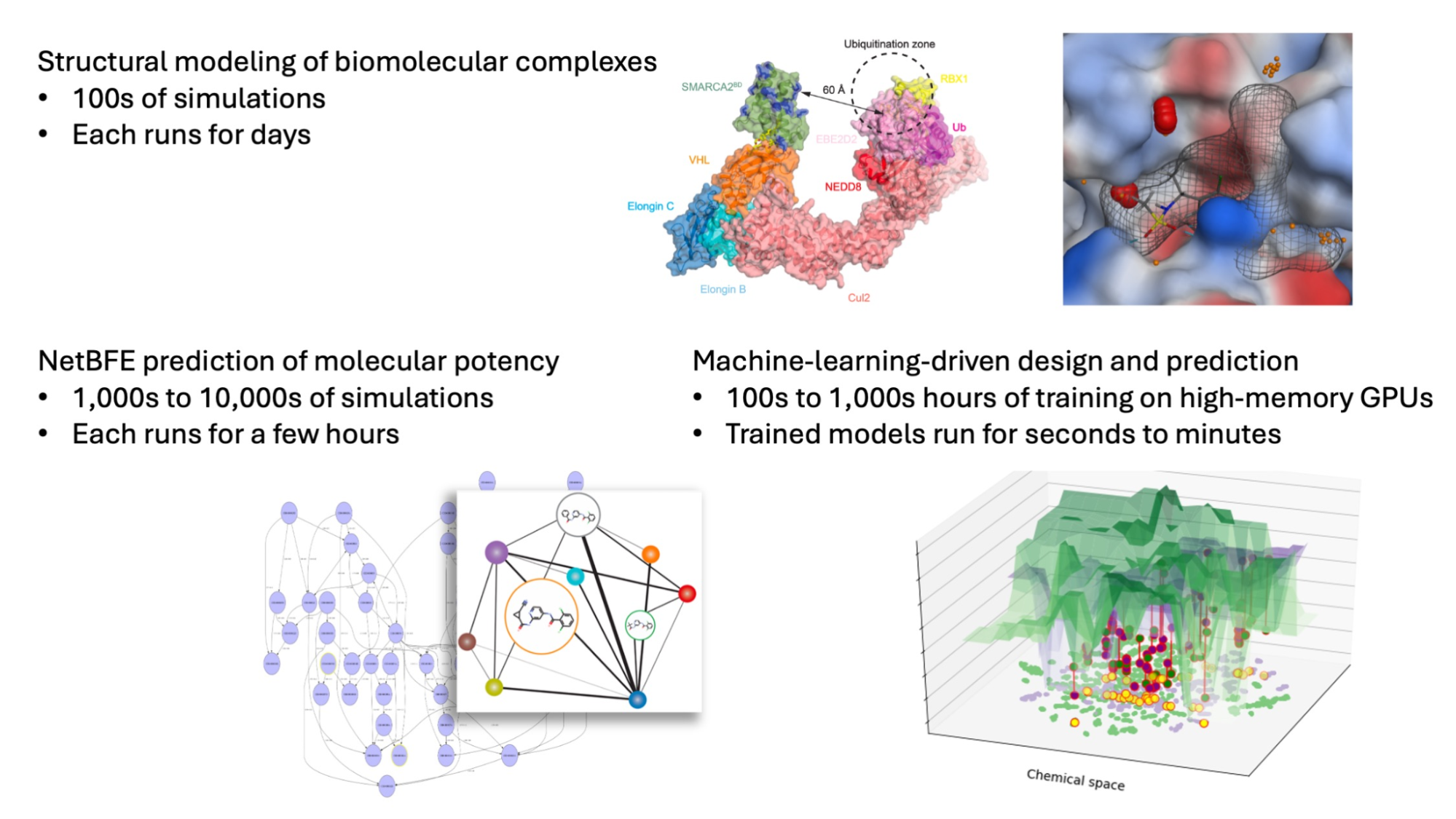
The computation-driven strategy for molecule discovery used by Atommap
Atommap molecular engineering platform leverages high-performance computing to identify new therapeutic compounds against targets that were previously unreachable through innovative processes. This approach expedites, reduces costs, and increases the likelihood of success in the process.
Atommap platform has significantly cut the time (by more than half) and cost (by 80%) of molecular discovery in previous projects. For instance, it significantly sped up the search for new compounds that degrade highly valuable oncological targets and was crucial in moving a molecule against a difficult therapeutic target to the clinical trial in 17 months (NCT04609579) (Mostofian et al. 2023).
Atommap does this through:
Complicated conformational dynamics of the protein target and its interactions with drug compounds and other biomolecules are revealed by sophisticated molecular dynamics (MD) simulations. They determine the target protein’s dynamics-function link, which is crucial for determining the most effective drug molecule mechanism of action.
Models that generate new molecules by counting them. Google Cloud models start with a three-dimensional blueprint of a drug molecule’s interaction with its target and then computationally generate thousands to hundreds of thousands of new virtual molecules that satisfy both favorable drug-like properties and the desired interactions.
Prediction models that are ML-enhanced and based on physics that can predict molecule potencies and other attributes with accuracy. The computer evaluation of each molecular design takes into account its drug-likeness, target-binding affinity, and effects on the target. As a result, Cloud may investigate many times more compounds than can be synthesized and tested in a lab. They can also carry out several rounds of design while that wait for the results of frequently drawn-out experiments, which shortens turnaround times and raises the likelihood of success.
Molecular Discovery as a Service and Computation as a Service
In order to have a meaningful and widespread influence on drug development, Atommap must combine its extensive knowledge of molecular discovery with outside knowledge of target identification, clinical trials, and commercialization.
They establish partnerships through two methods: Molecular development as a Service (MDaaS, pronounced Midas) and Computation as a Service (CaaS). Google cloud computation-driven molecular engineering platform is easily and affordably accessible to all drug development organizations.
Atommap pay-as-you-go cloud-as-a-service (CaaS) approach allows any discovery project to test their computational tools at a small and reasonable scale without committing a large amount of money, as opposed to selling software subscriptions. While not all projects can be solved computationally, the majority can. With this method, any drug development effort can swiftly and affordably introduce the necessary computations with observable effects, then scale them up to maximise their benefits.
Google cloud MDaaS cooperation enables drug hunters looking to turn their biology and clinical concepts into therapeutic candidates to swiftly find powerful compounds with unique intellectual property for clinical trials. From the first molecule (initial hits) to the final molecule (developing candidates), Atommap completes the molecular discovery project, freeing up partners to concentrate on biology and clinical validation.
How to construct a Google Cloud elastic supercomputer
Cloud computing platform was moved from their internal cluster to a hybrid environment that incorporates Google Cloud over the course of multiple steps.
Slurm
Slurm was used by numerous workflows in their platform to handle computing activities. They used Google’s open-source Cloud HPC Toolkit to create a cloud-based Slurm cluster in order to move to Google Cloud. A command line utility called Cloud HPC Toolkit makes it simple to set up safe, networked cloud HPC systems. They immediately built up computing workloads for Google cloud discovery projects using their Slurm-native tooling after this Slurm cluster was up and operating in a matter of minutes.
Their DevOps department naturally integrates into best practices with the help of Cloud HPC Toolkit. Google compute clusters are specified as “blueprints” inside of YAML files, which enable us to easily and transparently configure particular features of different Google Cloud products. The Toolkit converts blueprints into input scripts that are run by Terraform, an industry-standard tool from Hashicorp that defines “infrastructure-as-code” that can be version-controlled, committed, and reviewed.
Google Cloud also specified their compute machine image in the blueprint using a startup script that works with Hashicorp’s Packer. This made it simple for us to “bake in” the programmes that are usually required for Google cloud jobs, such conda, Docker, and Docker container images that supply dependencies like PyTorch, AMBER, and OpenMM.
Comparable in accessibility and ease of use to previous Slurm systems they have utilized is the deployed Slurm cloud system. They only pay for what Google Cloud use because the computing nodes are only deployed when requested and spun down when completed; the head and controller nodes, from which they log in and deploy, are the only persistent nodes.
Batch
The cloud-native Google Batch allows us much more freedom in their access to the computational resources when compared to Slurm. Batch can be used to plan cloud resources for lengthy scientific computing workloads because it is a managed cloud job-scheduling service. Batch builds up virtual machines that mount NFS stores or Google Cloud Storage buckets with ease. The latter is especially handy as an output directory for their long-running simulations, as it can house multi-gigabyte MD trajectories.
SURF-Submit, Upload, Run, Fetch
In the majority of computational procedures, a common pattern has surfaced. The sequence and structures of the target protein, a list of small compounds along with their valence and three-dimensional structures, the simulation and model parameters are the first set of sophisticated input files that are included in each task. Second, even on the fastest computers, the majority of computing tasks take hours or even days to complete.
Thirdly, the computer tasks generate output datasets that are subject to several analysis and have a sizable volume.

